The study of intricate molecular regulation has spanned several decades and involves depicting interactions among molecules in the form of networks. Within these networks, nodes represent objects of interest such as genes, transcription factors (TFs), and proteins. Consequently, numerous molecular networks have emerged, including gene regulatory networks (GRNs), protein-protein networks, and signal transduction networks. The recent availability of omics data collected over time has enabled the deduction of causal relationships between these nodes.
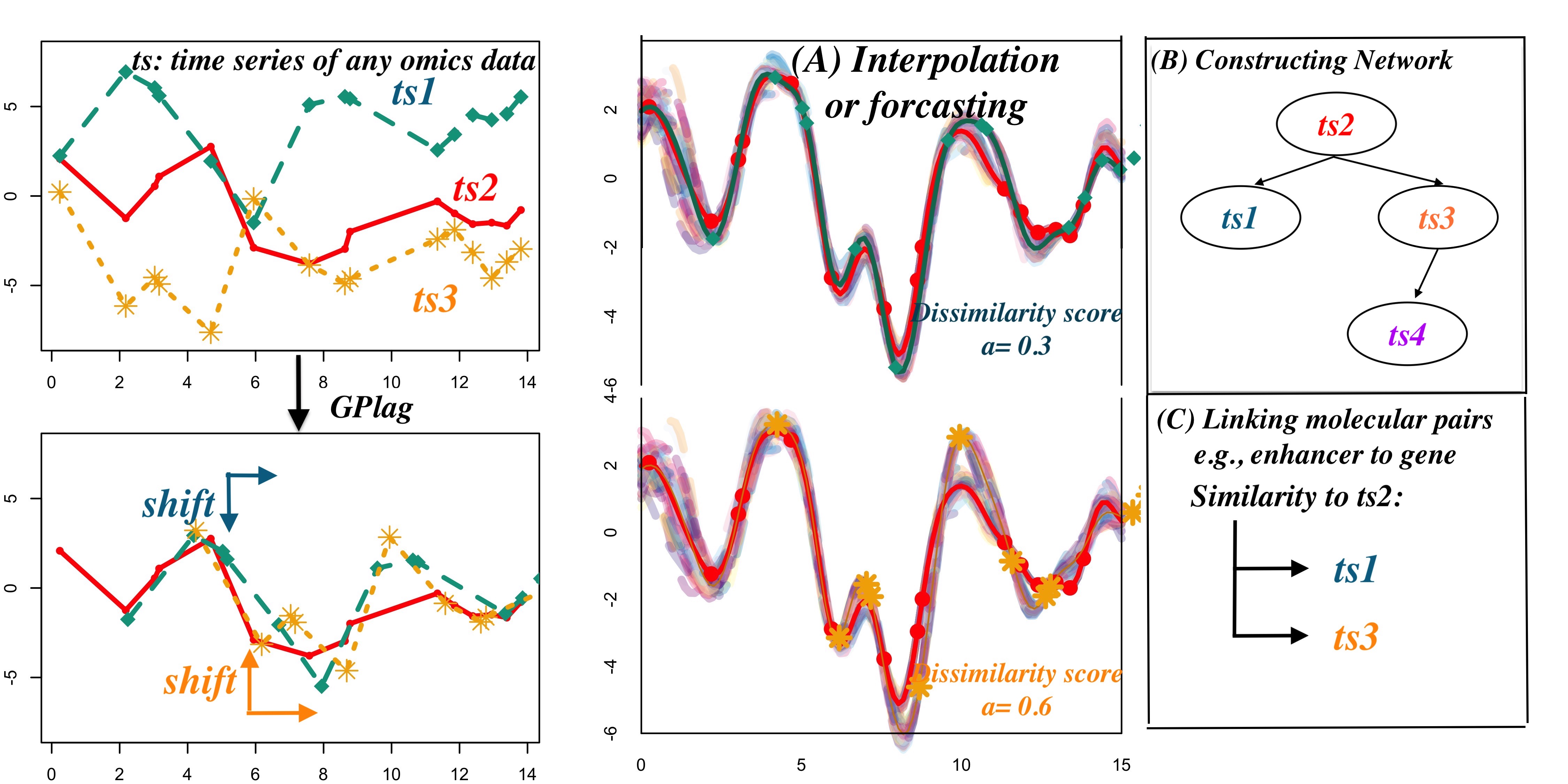
we have developed a model grounded in Gaussian processes (GPs). We address these challenges by developing a novel cross-covariance approach within the GP framework, incorporating interpretable kernel parameters. Our method is supported by a series of theoretical proofs that validate the effectiveness of our proposed kernels and demonstrate the identifiability of kernel parameters. This means that the theory guarantees the recovery of the true values of two essential kernel parameters, which measure the dissimilarity (denoted as a) and the time lag (denoted as s) between time series through estimation. Furthermore, GPlag operates on pairs of molecules at each step, enabling parallelization and significantly improving the efficiency of network inference, even when dealing with millions of potential edges. In summary, GPlag offers biological interpretability by providing information about the direction (sign), time lag, and the strength of the causal relationship between pairs of molecules. This information can be applied to various tasks, such as interpolation, network construction, and linking regulatory pairs

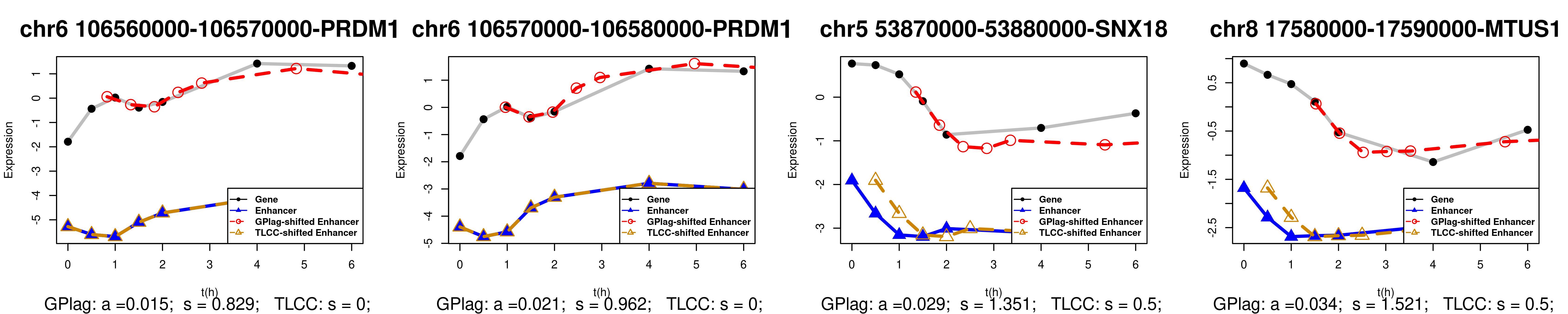
We validate our link between enhancer and gene using Hi-C (High-throughput Chromosome Conformation Capture): The candidate enhancer-gene pairs identified by our proposed method exhibit significantly higher Hi-C values compared to non-candidate pairs. In contrast, other state-of-the-art methods, constrained by issues such as sparsity and non-uniformity, fail to deliver significant results. Additionally, we utilize the causal GRN inferred by GPlag from time-series transcriptomics data to explore the relationships between immune genes and metabolic genes following macrophage stimulation