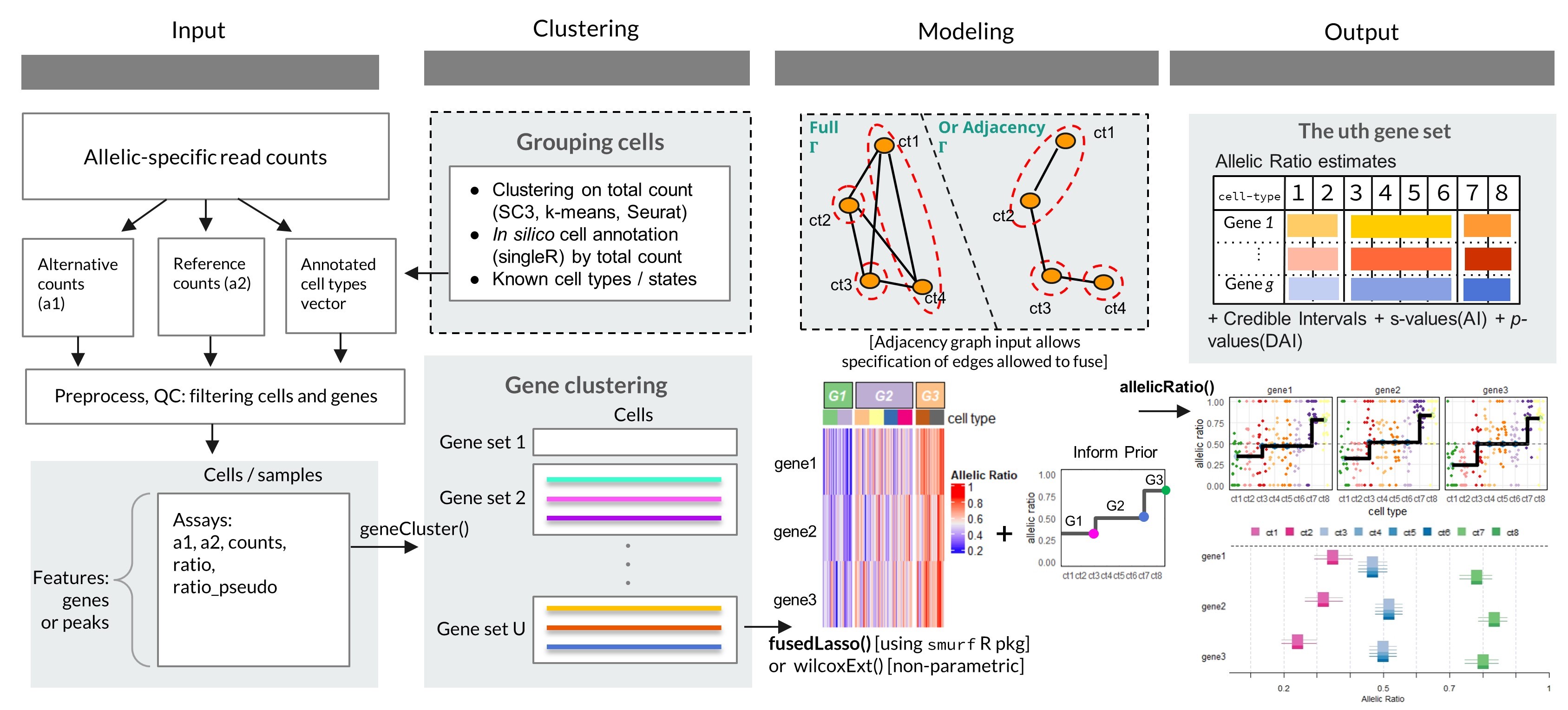
Single-cell RNA-seq of F1 crosses enables measurement of context-specific allelic expression (AE), where the cell type or cell stage can be taken as the context that influences cis-genetic regulation. Spatially resolved or time course allelic datasets offer another such example. airpart provides discrete grouping of cell types, providing interpretability to the fitted models. The groups provided by the partition step can help to generate hypotheses of CTS cis-regulatory mechanisms. For example, cell types within the same group may share a common mechanism of cis-regulation, such as a common set of expressed transcription factors and active regulatory elements harboring genetic variation. Additionally, airpart can be used to model continuous gradients of cis-regulatory effects on cells or samples.

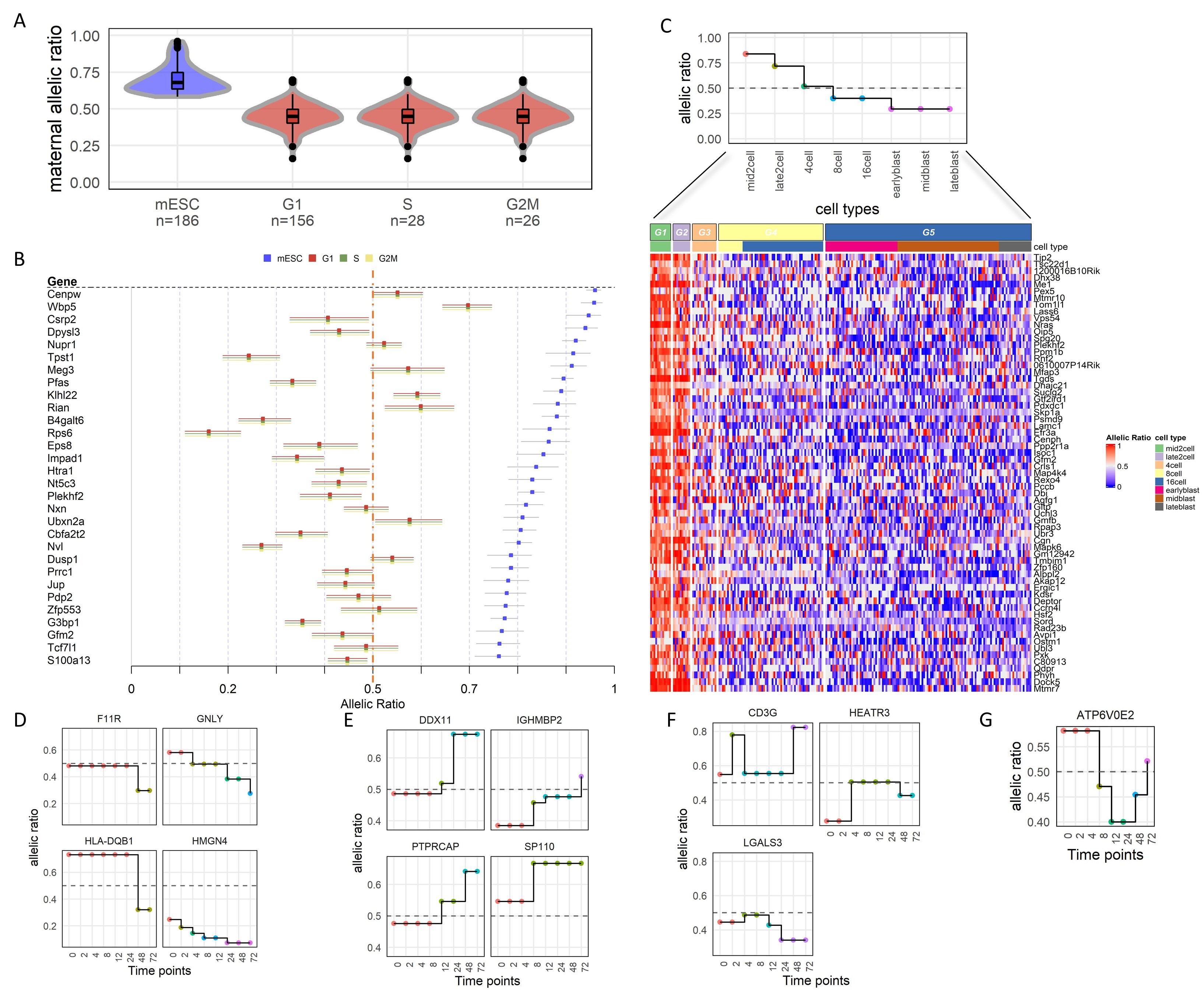
airpart partitioning of the time series by allelic ratio revealed four types of patterns (decreasing, increasing, up-peak and downpeak) as shown in Figure 3(D–G), respectively. As in the original study, we also observed the dominant allele could switch over the time course, or bi-allelically expressed genes could switch to dominant by one or the other allele. While the original paper used logistic regression with polynomial terms for time within each individual, we recovered similar DAI trends for many autoimmune genes, such as GNLY and DDX11. Overall, airpart successfully captured the DAI patterns seen across T-cell activation
Airpart: interpretable statistical models for analyzing allelic imbalance in single-cell datasets
Paper Published in Bioinformatics
We provided a tutorial for running airpart in details, with all kinds of visualization plots provided. Package Website: airpart Tutorial