Page Not Found
Page not found. Your pixels are in another canvas.
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
Page not found. Your pixels are in another canvas.
About me
This is a page not in th emain menu
Published:
This post will show up by default. To disable scheduling of future posts, edit config.yml and set future: false.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Published in Journal 1, 2009
This paper is about the number 1. The number 2 is left for future work.
Recommended citation: Your Name, You. (2009). "Paper Title Number 1." Journal 1. 1(1). http://academicpages.github.io/files/paper1.pdf
Published in Journal 1, 2010
This paper is about the number 2. The number 3 is left for future work.
Recommended citation: Your Name, You. (2010). "Paper Title Number 2." Journal 1. 1(2). http://academicpages.github.io/files/paper2.pdf
Published in Journal 1, 2015
This paper is about the number 3. The number 4 is left for future work.
Recommended citation: Your Name, You. (2015). "Paper Title Number 3." Journal 1. 1(3). http://academicpages.github.io/files/paper3.pdf
Deciphering regulation network or linking regulatory pairs in time-series omics data with consideration of time lag
A hierarchical Bayesian modeling to estimate cell type specific allelic imbalance per gene in single-cell RNA-seq or dynamic allelic imbalance per gene in bulk RNA-seq.
Convolutational neural network for predicting guide RNA effects in CRISPR epigenome editing experiments 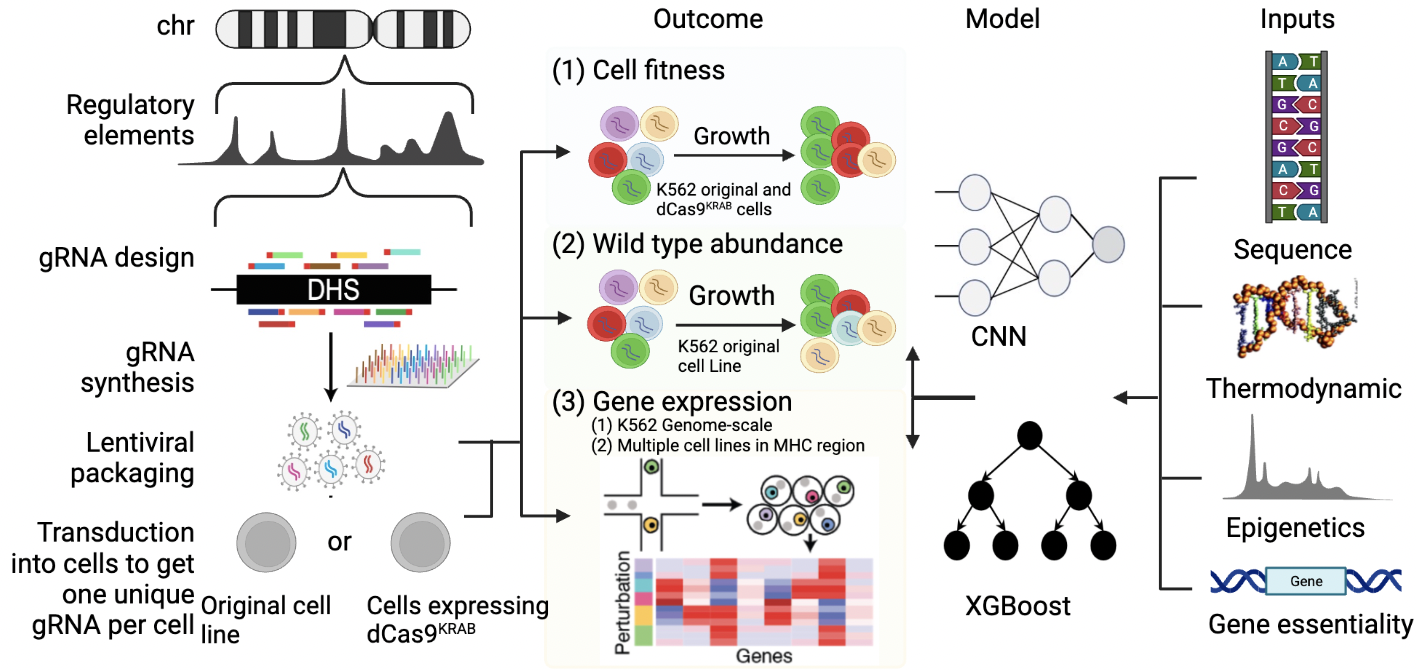
Generation of null ranges via bootstrapping or covariate matching.
Published:
This is a description of your talk, which is a markdown files that can be all markdown-ified like any other post. Yay markdown!
Published:
This is a description of your conference proceedings talk, note the different field in type. You can put anything in this field.